Docker based RStudio & PostgreSQL
This is part one of the two part post related to Docker, PostgreSQL databases and Anomaly data-sets.
Background
In recent LinkedIn posts (original and Rami’s repost) and tweet, I asked the internet for their favorite datasets for anomaly detection problems, particularly in the time-series domain. I got lots of responses, and now have a massive amount of data to play with, thank you folks who responded.
To play with these data, I wanted a better solution than keeping data as CSV files, qs objects, or in R Packages. I wanted a database to store the raw input data, my processed data and algorithm results. While I’ve setup databases in the past the traditional way, this time I wanted the entire codebase to be portable & reproducible.
As a result, I setup everything using Docker and it works seemlessly. If you’d like to learn how to do so, follow along my next posts:
- Part I - (this post) Will teach you how to setup a simple reproducible Docker based workflow for a personal PostgreSQL Database + RStudio for code development
- Part II - (next post) Will be the ETL pipelines for anomaly data
Why should you read this?
At the end of this tutorial, you’ll be able to rapidly setup a Docker based personal1 PostgreSQL database. You will learn how to quickly deploy PostgreSQL & RStudio using docker-compose. You will be able to access the database in R and begin development immediately. And most importantly, the whole process will be fully reproducible as you inherit the benefits of setup scripts and Docker.
This tutorial assumes you’re familiar with Docker and RStudio in a Docker environment. If you’re not, I recommend reading Reproducible Work in R first.
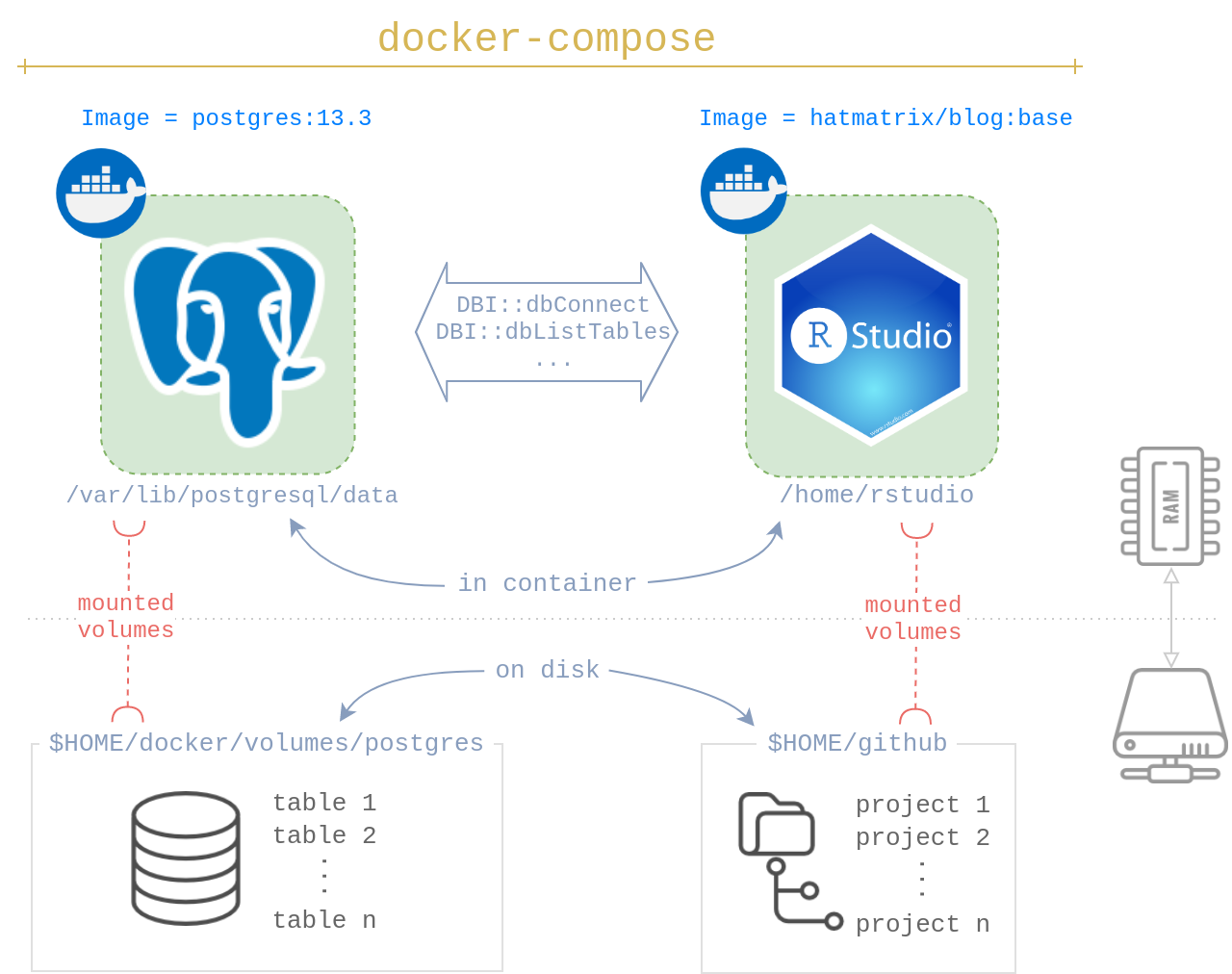
Overview
You will be launching two Docker images:
- A PostgreSQL image. I choose
postgres:13.3 - An RStudio image. I choose
hatmatrix/blog:base2
To permanently store your data beyond the life of the containers, you will mount two volumes, one for each container. I chose:
- For PostgreSQL:
$HOME/docker/volumes/postgres - For R Projects:
$HOME/github
None of these paths, except for the in-container PostgreSQL are special; you can customize the others to your liking. By default, postgres:13.3 expects the database to be at /var/lib/postgresql/data. If you choose another database, modify this accordingly.
I use docker-compose to launch both PostgreSQL and RStudio services together. It’s convenient while also ensuring the PostgreSQL service runs first followed by RStudio. It’s easy to start or stop all the services using just a few commands.
First Time Setup
You need to run these steps the first time you’re setting up the PostgreSQL database. I’ve stored these steps in 00-postgres-init.sh.
1 - Directory Setup
You need a local directory to store the PostgreSQL database in. Lines 3-10 take care of this for you.
# create directory if does not exist
if [ -d "$HOME/docker/volumes/postgres" ]
then
echo "Directory $HOME/docker/volumes/postgres exists."
else
echo "Error: Directory $HOME/docker/volumes/postgres does not exists."
mkdir -p $HOME/docker/volumes/postgres
fi2 - PostgreSQL Setup
Now it’s time to setup the database. You need two steps at a minimum to get started:
- A new ‘role’ (akin to a login) with rights to create new databases.
- At least one database to work in. In my script, I’m making two:
workandanomaly.
To manipulate the database, you need a PostgreSQL server running to process the psql commands. You’ll launch one using docker run. You need the correct volume mounted using -v. Next, we create the role and databases by piping psql commands into docker exec. Then, we stop the container.
# launch the postgres image called 'post_setup',
# attach it to the local volume
docker run --rm --name post_setup \
-e POSTGRES_USER=postgres \
-e POSTGRES_PASSWORD=docker \
-d \
-p 5432:5432 \
-v $HOME/docker/volumes/postgres:/var/lib/postgresql/data \
postgres:13.3
# create a new role, and two databases
echo "CREATE ROLE rahul WITH PASSWORD 'pass' CREATEDB LOGIN;
CREATE DATABASE work; CREATE DATABASE anomaly;" | \
docker exec -i post_setup \
psql -U postgres
# stop the docker container
docker stop post_setupIn summary, now I have a PostgresSQL database:
- stored at
$HOME/docker/volumes/postgres - with a new role
rahuland passwordpass - with 2 databases:
workandanomaly
Daily Workflow
tldr: How do you get going?
- Store
docker-compose.ymlin a local directory - Modify it if you’ve changed my chosen images/directories
- In shell, run
docker-compose up -d
protip: to launch a browser (firefox for me) directly into RStudio as well, run this command in the directory where you have docker-compose.yml:
docker-compose up -d; firefox localhost:8787pro-protip: save an alias and generalize the command. The -f arg instructs docker-compose which file you’d like to use. Now engage can be run from anywhere in the system.
alias engage='docker-compose \
-f $HOME/github/docker/docker-compose.yml \
up -d; firefox localhost:8787' The breakdown
What’s in docker-compose.yml? We’re creating two services, one called db and the other rstudio.4
Let’s look at db first. Most of the arguments will look familiar if you’re familiar with docker run args. What’s new here is the restart: unless-stopped arg which tells Docker to only start PostgreSQL if it’s currently stopped.
version: "3.3"
services:
db:
image: postgres:13.3
restart: unless-stopped
environment:
POSTGRES_DB: "anomaly"
POSTGRES_USER: "rahul"
POSTGRES_PASSWORD: "pass"
ports:
- "5432:5432"
volumes:
- type: "bind"
source: "$HOME/docker/volumes/postgres"
target: "/var/lib/postgresql/data"The 2nd service is rstudio. Apart from the typical args, the interesting arg here is depends_on which tells Docker to only run this image after the database is up and running. Fantastic!
rstudio:
image: hatmatrix/blog:base
ports:
- "8787:8787"
- "3838:3838"
environment:
DISABLE_AUTH: "true"
volumes:
- type: "bind"
source: "$HOME/github"
target: "/home/rstudio"
depends_on:
- "db"Connecting via R
Use postgres.R to test your connection. Run your DBI:: commands you would normally, except for one key difference.
While making the connection, make sure the name of the host is the name of the database service you’ve chosen in docker-compose.yml. (Outside docker, you would have typically used localhost to connect to a local PostgreSQL server).
con <- DBI::dbConnect(
drv = RPostgres::Postgres(),
dbname = "anomaly",
host = "db", # this needs to be the name of the postgres service
# (line 3 in docker-compose.yml)
user = "rahul",
password = "pass",
port = 5432
)That’s it! You’re off to the races now. Use the DB as you normally would using {DBI}.
con %>% DBI::dbListTables()
con %>% dplyr::tbl("table_name")To Stop Services
You have two options here:
docker-compose stopwill stop the services, which you can restart usingdocker-compose start.docker-compose downwill and remove containers as well. Rundocker-compose upto get going once again.
Footnotes
I harp on ‘personal’ given I’m not setting up appropriate roles, auth etc needed in a work environment. But, it’s good enough for my personal use.↩︎
This is my own image based off of
rocker/rstudio↩︎- DB icon by Pixel perfectFolder icon by Icongeek26RAM icon by FreepikStorage icon by Smashicons↩︎
These are just labels, you can call them what you’d like↩︎